|
Hybridization probe / Molecular Probes / FISH / CISH
Gene
/ Clinical Panels / Disease / Region / Fusion / Break Apart / Controls / Human BAC /
Mouse BAC / Custom / Rapid Hybridzation Buffer / Amplification / Controls Probes
https://en.wikipedia.org/wiki/Hybridization_probe
https://en.wikipedia.org/wiki/Fluorescence_in_situ_hybridization
https://en.wikipedia.org/wiki/Chromogenic_in_situ_hybridization
https://en.wikipedia.org/wiki/Immunohistochemistry
Probes – RNA and DNA
RNA probes can be designed for any gene or any sequence within a gene for
visualization of mRNA,[3][4][5]
lncRNA[6][7][8] and
miRNA in tissues and cells. FISH is used by examining the cellular reproduction
cycle, specifically interphase of the nuclei for any chromosomal abnormalities.[9] FISH
allows the analysis of a large series of archival cases much easier to identify
the pinpointed chromosome by creating a probe with an artificial chromosomal
foundation that will attract similar chromosomes.[9] The
hybridization signals for each probe when a nucleic abnormality is detected.[9] Each
probe for the detection of mRNA and lncRNA is composed of 20 oligonucleotide
pairs, each pair covering a space of 40–50 bp. For miRNA detection, the probes
use proprietary chemistry for specific detection of miRNA and cover the entire
miRNA sequence.....
|

ViewRNA detection of miR-133(green) and myogenin mRNA (red) in C2C12
differentiating cells. |
Probes are often derived from fragments of DNA that were isolated, purified, and
amplified for use in the Human Genome Project.
The size of the human genome is so large, compared to the length that could be
sequenced directly, that it was necessary to divide the genome into fragments.
(In the eventual analysis, these fragments were put into order by digesting a
copy of each fragment into still smaller fragments using sequence-specific
endonucleases, measuring the size of each small fragment using size-exclusion chromatography,
and using that information to determine where the large fragments overlapped one
another.) To preserve the fragments with their individual DNA sequences, the
fragments were added into a system of continually replicating bacteria
populations. Clonal populations of bacteria, each population maintaining a
single artificial chromosome, are stored in various laboratories around the
world. The artificial chromosomes (BAC)
can be grown, extracted, and labeled, in any lab containing a library. Genomic
libraries are often named after the institution in which they were developed. An
example being the RPCI-11 library, which is named after the Roswell Park Cancer
Institute in Buffalo NY. These fragments are on the order of 100 thousand
base-pairs, and are the basis for most FISH probes.....
https://en.wikipedia.org/wiki/Fluorescence_in_situ_hybridization#Probes_%E2%80%93_RNA_and_DNA
Preparation and hybridization process – RNA
Cells, circulating tumor cells (CTCs),
or formalin-fixed paraffin-embedded (FFPE) or frozen tissue sections are fixed,
then permeabilized to allow target accessibility. FISH has also been
successfully done on unfixed cells.[10] A
target-specific probe, composed of 20 oligonucleotide pairs, hybridizes to the
target RNA(s). Separate but compatible signal amplification systems enable the
multiplex assay (up to two targets per assay). Signal amplification is achieved
via series of sequential hybridization steps. At the end of the assay the tissue
samples are visualized under a fluorescence microscope.....
|
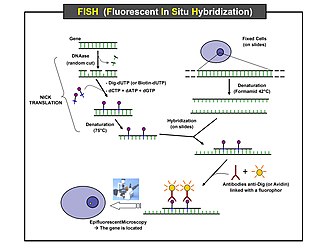
Scheme of the principle of the FISH Experiment to localize a gene in the
nucleus. |
Preparation and hybridization process – DNA
First, a probe is constructed. The probe must be large
enough to hybridize specifically with its target but not so large as to impede
the hybridization process. The probe is tagged directly with fluorophores, with targets for antibodies or with biotin. Tagging can be done in various ways, such as nick translation, or Polymerase chain reaction using
tagged nucleotides.
Then, an interphase or metaphase chromosome preparation is
produced. The chromosomes are firmly attached to a substrate, usually glass. Repetitive
DNA sequences must be blocked by adding short fragments of DNA to the sample.
The probe is then applied to the chromosome DNA and incubated for approximately
12 hours while hybridizing. Several wash steps remove all unhybridized or
partially hybridized probes. The results are then visualized and quantified
using a microscope that is capable of exciting the dye and recording images.
If the fluorescent signal is weak, amplification of the signal may be necessary
in order to exceed the detection threshold of the microscope. Fluorescent signal strength depends on
many factors such as probe labeling efficiency, the type of probe, and the type
of dye. Fluorescently tagged antibodies or streptavidin are bound to the dye
molecule. These secondary components are selected so that they have a strong
signal......
|
Chromogenic in situ hybridization (CISH)
is a cytogenetic technique that combines the chromogenic signal detection method
of immunohistochemistry (IHC) techniques
with in
situ hybridization.[1][2] It
was developed around the year 2000 as an alternative to fluorescence in
situ hybridization (FISH) for
detection of HER-2/neu oncogene amplification.[1] CISH
is similar to FISH in that they are both in
situ hybridization
techniques used to detect the presence or absence of specific regions of DNA.[1] However,
CISH is much more practical in diagnostic laboratories because it uses
bright-field microscopes rather than the more expensive and complicated
fluorescence microscopes used in FISH.[1][3]
.....
Probe design for CISH is very similar to that for FISH
with differences only in labelling and detection. FISH probes are generally
labelled with a variety of different fluorescent tags and can only be detected
under a fluorescence microscope,[4] whereas CISH probes are labelled with biotin or
digoxigenin [5] and can be detected using a bright-field microscope after
other treatment steps have been applied.[1]
CISH probes are approximately
20 nucleotides in length and are designed for DNA targets. They are
complementary to the targeted sequence and bind to it after a denaturation and
hybridization step. Only a few CISH probes are available commercially, so for
most applications they have to be extracted, amplified, sequenced, labelled and
mapped from bacterial
artificial chromosomes (BACs).[6] |

|
BACs were developed during the Human Genome Project as
it was necessary to isolate and amplify short fragments of human DNA for
sequencing purposes.[7] Nowadays,
BACs can be selected and positioned on the human genome using public databases
such as the UCSC Genome Browser.[6] This
ensures optimal complementarity and sequence specificity. DNA is extracted from
the BAC clones and amplified using a polymerase-based technique, such as
degenerate oligonucleotide primed (DOP)-PCR.[8] Next,
the clones are sequenced and their position on the genome is verified.[9]
Probe labelling can be carried out by using either random priming or nick translation to
incorporate biotin or digoxigenin.[10] |
|
|
Compared to FISH
https://en.wikipedia.org/wiki/Flow_cytometry#Measurable_parameters
CISH has some advantages over FISH in the reagents and
equipment it uses. As noted above, CISH is much cheaper and is easier to use
because it uses bright-field microscopes instead of fluorescence microscopes.[1][3] In addition,
the CISH reagents are more stable than the FISH reagents so it is possible to
store the samples and examine the same sample multiple times.[3][14] FISH reagents
fade over time due to photobleaching so a sample can only be examined once.[3][14] Apart from the
expensive fluorescence microscope, FISH also requires a high-resolution digital
camera to capture micrographs of the sample before the fluorescence fades.[14] Another
advantage of using bright-field microscopy is that the tissue or cell sample as
a whole can be visualized through CISH whereas cell morphology is difficult to
assess using fluorescence microscopy in FISH.[3][14]
|
|
CISH also differs from FISH in the probes that are used
as well as in the overall method. There are many different types of FISH probes
available, such as repeat probes, probes that detect specific genes or
telomeres, and probes that detect chromosomal abnormalities.[14] In contrast,
there is a limited variety of commercially available CISH probes, including
probes that bind the centromere of chromosomes 3, 7, 8, 9, 10, 11, 17, 18, X,
and Y as well as gene-specific probes for cancer-related genes, such as HER-2,
EGFR, MYC, and TOP2A.[14] Despite the
limited variety of available CISH probes, they are generally more cost-effective
than FISH probes.[14] With regard to
the overall method, FISH can be performed using direct labelling—fluorochromes
are attached to the probes—or indirect labelling—the probes are labelled with
biotin or digoxigenin which are then detected using fluorescently-labelled
streptavidin or antibodies, respectively.[14] CISH is
performed using indirect labelling in which antibodies or streptavidin are
conjugated to enzymes such as HRP or alkaline phosphatase
(AP).[14]
|
|
Compared to IHC
CISH and IHC are similar in that both are used for the
same purpose (mainly to detect HER-2/neu amplification) and they both use enzyme
reactions (HRP/AP) to measure amplification.[13] CISH and IHC are different in that IHC measures protein
expression whereas CISH measures DNA amplification.[13] This difference is particularly useful for HER-2/neu
because it has been found that gene amplification is of higher prognostic value
than protein expression.[17]
A disadvantage of IHC is that it is not possible to
identify false-negative and false-positive results.[17] In CISH, if there is no signal for the reference probe,
the assay has failed.
For low and high protein overexpression/gene
amplification, CISH and IHC show a concordance of over 86% and over 89%,
respectively.[18] It has been shown that monoclonal antibodies are
better than polyclonal antibodies for
detection in both IHC and CISH as they bind mor
|

|
especifically, which leads to a higher concordance
rate.[18]
For medium protein overexpression/gene amplification
concordance varies, but is higher when monoclonal antibodies are used than when
polyclonal antibodies are used.[18] The variable concordance is due to the fact that gene
amplification does not strictly correlate with protein expression and that tumor
heterogeneity can make it difficult to detect protein overexpression in a
tissue.[18] |

|
Medical applications
CISH is frequently applied to assess
Gene Amplification,
such as HER-2/neu status in breast cancer samples.[2][19] HER-2/neu
amplification is associated with higher mortality, higher recurrence rate, and
poor prognosis in breast cancer.[20] The monoclonal
antibody trastuzumab is a receptor blocker that
has been proven to be clinically very effective in HER-2/neu-overexpressing
tumors.[21] Therefore, it
is crucial to determine receptor status before starting cancer treatment.[21]
CISH is also used for detection of
Chromosomal
Rearrangements and fusions, such as the fusion of ALK tyrosine kinase domain
with the promoter and 5’ region of EML4 in lung cancer. ALK-positive tumors are
a clinically relevant subgroup as they can be very effectively treated with the
ALK inhibitor crizotinib.[22][23]
Apart from cancers, CISH has also been shown to be useful
in detecting human papillomavirus infections.[24]
|
Leading with Quality, Performance and Cost
Our partners, the
Empire Genomics, being a leading
professional developer and manufacturer dedicated for custom labeled Molecular
Probes for bioscience research and Clinical diagnostic applications, including
the
Fluorescence in situ hybridization (FISH)
and
Chromogenic in situ hybridization (CISH)
probes.

Logo of the HGP – the Vitruvian Man by Leonardo da Vinci. |
Empire Genomics
designs, manufactures and distributes more than one million clinical and
custom labeled
molecular probes, enabling hundreds of leading global clinical laboratories,
biotechnology organizations and academic institutions to advance biomarker
research, accelerate diagnoses and improve personalized treatment options for
patients battling cancer and other complex diseases.
The company was founded in 2006 by
Norma J. Nowak, PhD,
a prominent member of the
Human Genome
Project, to utilize innovative research started at the prestigious
Roswell Park Cancer Institute in Buffalo, New York.
The comprehensive product portfolio includes Fluorescence in Situ Hybridization
(FISH) and Chromogenic in Situ Hybridization (CISH) probes that are designed and
optimized for specific diseases, genes, and/or regions across the entire human
or mouse genomes... |
Probes – RNA and DNA
Extensive and mass amount of routine categories and customizable items are
available, ranging from probes for Gene, Clinical Panels, Disease, Region,
Fusion, Break Apart, Controls, Human BAC, and Mouse BAC, and Rapid Hybridzation
Buffer, etc.
|